- [GENOME MEDICINE] Integrative analysis of spatial and single-cell transcriptome data from human pancreatic cancer reveals an int
- 관리자 |
- 2024-07-24 09:20:45|
- 252
- 2024-07-24 09:20:45|
[Title]
Integrative analysis of spatial and single-cell transcriptome data from human pancreatic cancer reveals an intermediate cancer cell population associated with poor prognosis
[Author]
1 Graduate School of Medical Science and Engineering, Korea Advanced Institute of Science and Technology, 291 Daehak‑Ro, Yuseong‑Gu, Daejeon 34141, Republic of Korea.
2 Division of Gastroenterology, Department of Internal Medicine, Severance Hospital, Yonsei University College of Medicine, 50‑1 Yonsei‑Ro, Seodaemun‑Gu, Seoul 03722, Republic of Korea.
3 Department of Pharmacy and Yonsei Institute of Pharmaceutical Sciences, College of Pharmacy, Yonsei University, Incheon, Republic of Korea.
4 Department of Internal Medicine, Graduate School of Yonsei University, Seoul, Republic of Korea.
5 Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, Yonsei Cancer Center, Yonsei University College of Medicine, Pancreatobiliary Cancer Center, Severance Hospital, 50‑1 Yonsei‑Ro, Seodaemun‑Gu, Seoul 03722, Republic of Korea.
6 Pancreatobiliary Cancer Center, Yonsei Cancer Center, Severance Hospital, Seoul, Republic of Korea.
[Journal]
Genome Medicine volume 16, Article number: 20 (2024)
[Abstract]
Background
Recent studies using single-cell transcriptomic analysis have reported several distinct clusters of neoplastic epithelial cells and cancer-associated fibroblasts in the pancreatic cancer tumor microenvironment. However, their molecular characteristics and biological significance have not been clearly elucidated due to intra- and inter-tumoral heterogeneity.
Methods
We performed single-cell RNA sequencing using enriched non-immune cell populations from 17 pancreatic tumor tissues (16 pancreatic cancer and one high-grade dysplasia) and generated paired spatial transcriptomic data from seven patient samples.
Results
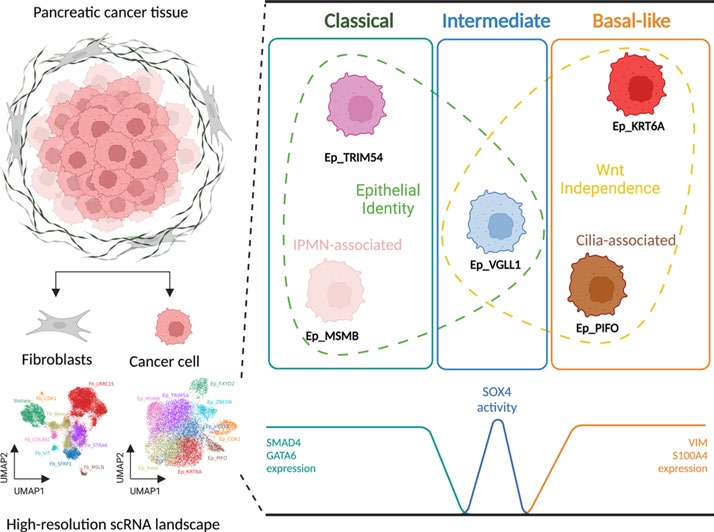
We identified five distinct functional subclusters of pancreatic cancer cells and six distinct cancer-associated fibroblast subclusters. We deeply profiled their characteristics, and we found that these subclusters successfully deconvoluted most of the features suggested in bulk transcriptome analysis of pancreatic cancer. Among those subclusters, we identified a novel cancer cell subcluster, Ep_VGLL1, showing intermediate characteristics between the extremities of basal-like and classical dichotomy, despite its prognostic value. Molecular features of Ep_VGLL1 suggest its transitional properties between basal-like and classical subtypes, which is supported by spatial transcriptomic data.
Conclusions
This integrative analysis not only provides a comprehensive landscape of pancreatic cancer and fibroblast population, but also suggests a novel insight to the dynamic states of pancreatic cancer cells and unveils potential therapeutic targets.
Graphical Abstract